In their 2011 Nature Reviews Drug Discovery paper “A call to reform the taxonomy of human disease” *, Ismail Kola and John Bell formulate an impressive plea for “A coordinated effort to incorporate advances in the understanding of the molecular and genomic variations in common diseases, such as hypertension, into their diagnosis and treatment could transform drug development and medicine.”
The two authors emphasize the need for a re-classification of diseases according to mechanisms that contribute to the aetiology of a disease at the molecular (“omics-“) level rather than the current phenotypic approach. They point out that several diseases that appear to be distinct at the clinical phenotype level may actually go back to one shared disease mechanism, or that diseases classified under one concept are in fact phenotypically similar manifestations of different, diverse disease mechanisms.
Kola and Bell underline that the concept of a mechanism-based taxonomy would also address the challenge of subgroup-specific or even individualised medicine, as an in-depth knowledge about the mechanisms underlying a disease inherently provides the basis for strategies to treat that disease in a dedicated, mechanism-specific way.
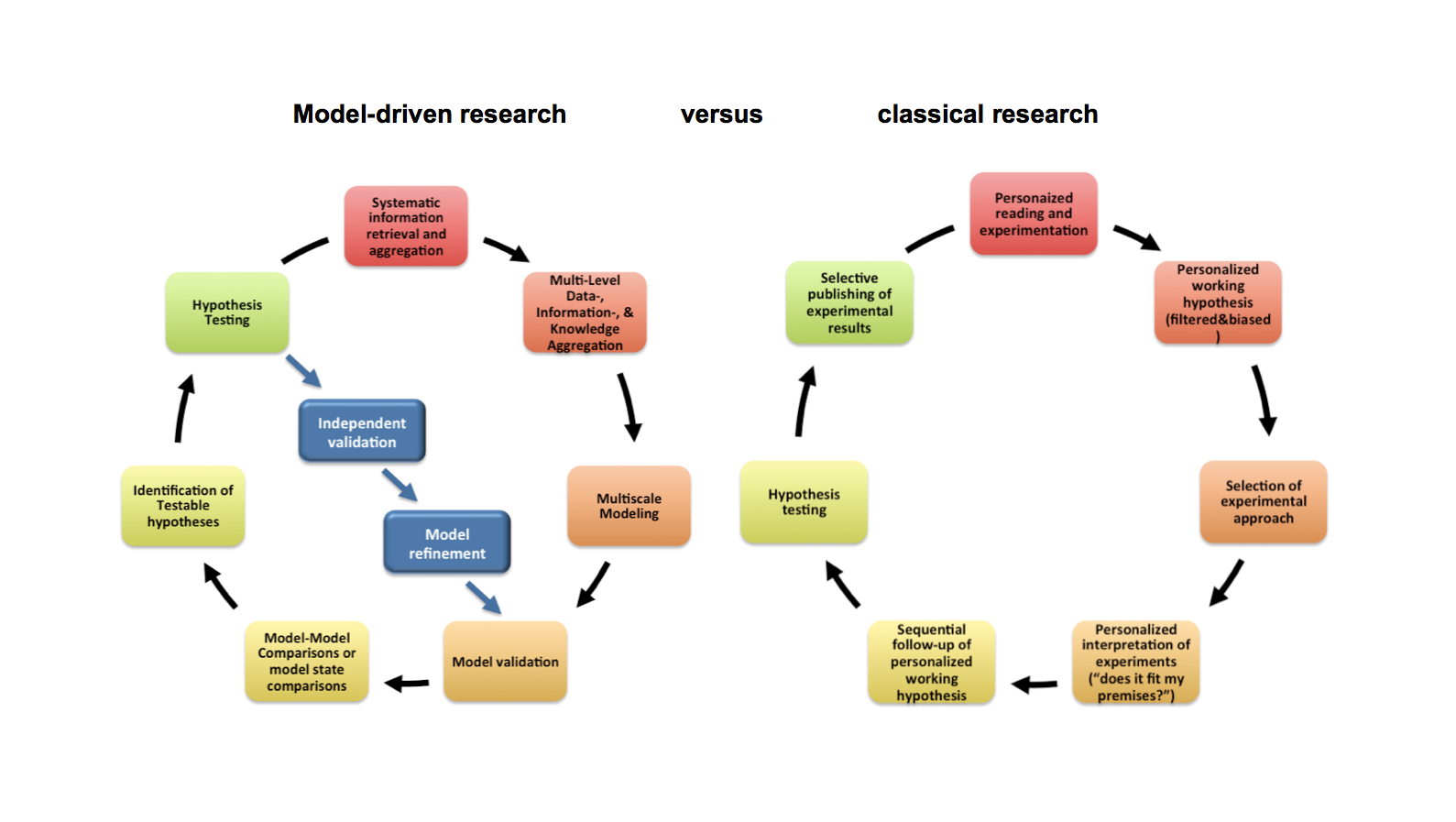
AETIONOMY is a response to the challenge they laid down and is a knoweldge generation approach which can accommodate the specific data types and data structures for a broad variety of indication areas whilst retaining the capacity to construct disease-specific taxonomies based on the underlying disease mechanisms. It is part of a larger, coordinated effort to establish a mechanism-based taxonomy which ultimately will result in improved diagnostic and therapeutic procedures.
 Organising Knowledge about Neurodegenerative Disease Mechanisms for the Improvement of Drug Development and Therapy
Organising Knowledge about Neurodegenerative Disease Mechanisms for the Improvement of Drug Development and Therapy